Revealing Stochastic Switches in Bacteria
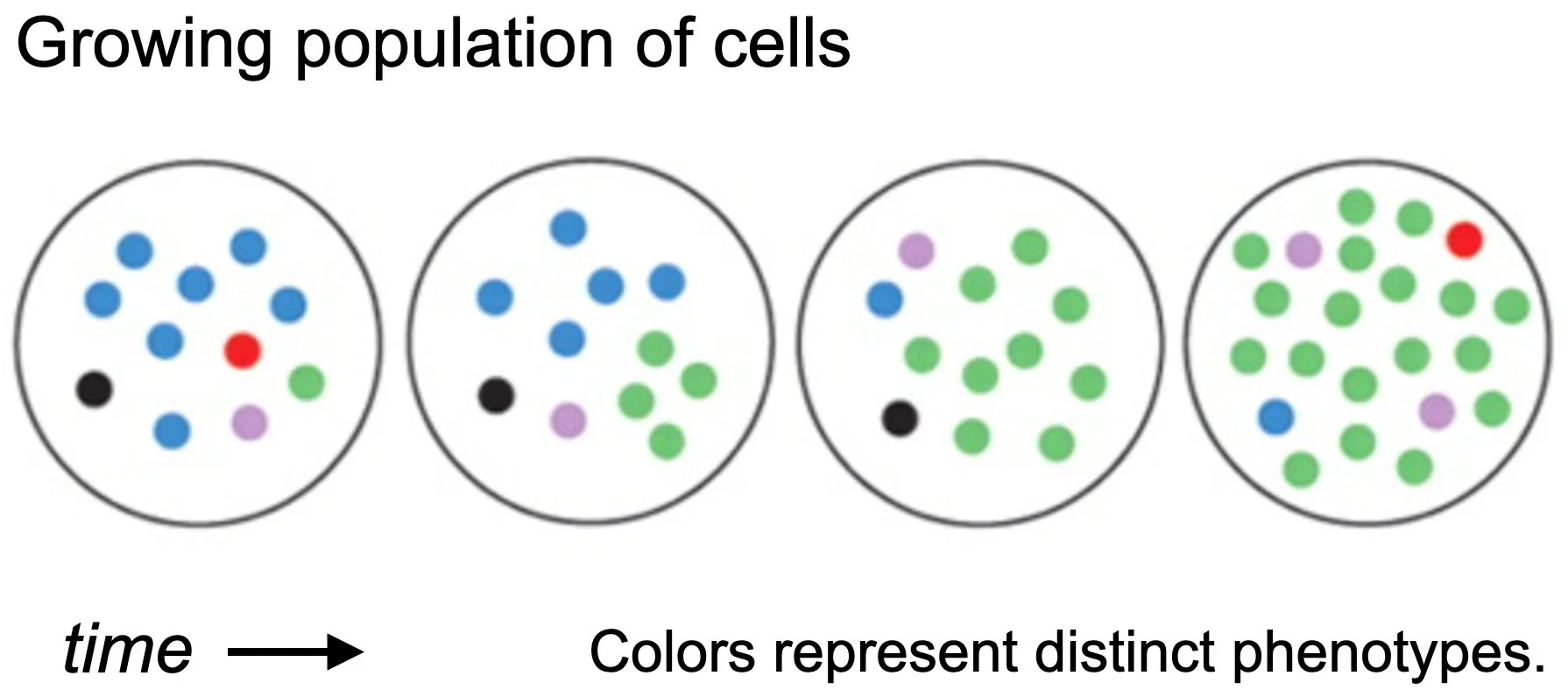
Our overarching goal is to understand how and why stochastic switches evolve.
We develop experimental and theoretical approaches to address this question in general, and apply these to specific systems of biomedical relevance, including emergence of antibiotic resistance, the role of bottlenecks in host-pathogen interactions, maintenance of immune defenses, and bacterial physiology in fluctuating environments.Stochastic switches are molecular mechanisms that enable cells to change their phenotype spontaneously and to maintain their phenotype heritably without sensing the environment. In bacteria, stochastic switches are an important class of survival mechanism, which allows a large population of cells to maintain subpopulations pre-adapted to different stressful environments.
Emergence of antibiotic resistance from single cells
Stochastic switches that control antibiotic resistance genes are prevalent in bacterial genomes. We developed a model system to track the emergence and dynamics of antibiotic resistance in single cells. By exposing cells to antibiotic pulses of different frequencies, we determine how environmental fluctuations and cellular physiology can mitigate the ability of resistance to sweep in a population.
Dynamics of antibiotic persistence
We collaborate with the Wakamoto Lab at the University of Tokyo’s Research Center for Complex Systems Biology to study the mechanisms by which antibiotic persistence operates in Escherichia coli. The Wakamoto Lab has developed custom microfluidic devices that are etched directly in glass, enabling extremely stable single cell observation over long timescales. Recent experiments have revealed the persistence dynamics of wild-type E. coli in exponential phase, which were previously inaccessible to direct observation due to their extremely low frequencies.
Impact of bottlenecks on evolution of stochastic switches in host-pathogen interaction
Pathogenic bacteria use stochastic switches to generate combinatorial diversity in their surface antigens, which enables evasion of host immune responses. Strong bottlenecks during host-to-host transmission impose selective constraints on the evolution of stochastic switching rates. Using mathematical modeling, we demonstrate that bottlenecks not only select for high switching rates, but also significantly speed up the evolutionary dynamics of stochastic switches.